Design, build, work
A physics engine for cell engineering
Welcome to The Century of Biology! This newsletter explores data, companies, and ideas from the frontier of biology. You can subscribe for free to have the next post delivered to your inbox:
Enjoy! 🧬
We can program cells to produce drugs, perform logic operations, and attack tumors.
What does it take to write a new cellular program? Trial and error, of course. Biology is complicated, and we shouldn’t ever expect our designs to work the first time. Individual genes interact in unexpected ways—the consequence of working with emergent systems. The best we can do is to explore as many designs as possible in parallel, accelerating the unavoidable Design, Build, Test, Learn engineering cycle.
What if that wasn’t true? In other words, what if we could…
Design, Build, Work.
When an architect designs a new building, we don’t test millions of prototypes in parallel. The first building just works. We’ve routinized the process of macroscopic construction.
This is also true for complex engineering domains. The Boeing 777 was the first plane designed entirely using Computer-Aided Design (CAD) software.1 Small things can also be designed with software, even really small things. The complex digital circuitry in a modern microprocessor is described in a high-level computer language before it is compiled into physical semiconductor-based circuits.
Cells are a different story. New designs often require years of experimentation and tinkering in the lab. Nothing ever works the first time.
If we can achieve this level of precision for aeronautical and electrical engineering, why does the design process for biology remain so empirical? To start, we don’t know what everything in a cell does. In 2016, researchers from the J. Craig Venter Institute published the first description of a minimal bacterial genome containing only the genes that are essential for life. Out of the 473 genes in this stripped-down genome, we don’t know what 149 of them do.2
To make matters worse, it’s incredibly hard to model interactions between genes reliably. Imagine trying to compose building blocks that you don’t understand very well. Our incomplete understanding of the physics of cells complicates things even further. We can’t model the environment in which the action happens.
Needless to say, bioengineers have a hard job. Here, we’re going to explore an attempt to make things easier. A new paper led by Akshay J. Maheshwari and Jonathan Calles from the Endy Lab at Stanford described a system for using a “physics engine” for cells to predictably engineer biological behaviors. We’ll examine how this works and why it might be a promising direction for the routinization of cellular-scale synthetic biology.
Colloidal dreams
When I arrived at Stanford, I knew I wanted to meet Drew Endy. Having been described as “synthetic biology’s most compelling evangelist,” few people have had a greater impact on the dreams of bioengineers around the world. Consider the Atoms are Local thesis, where Endy emphasizes the profound capacity for biological systems to produce complex matter all over the planet:
The leaves on a tree don’t come from a factory and then get shipped to where the tree is going to be and taped and stapled to the twigs and branches. The photons and molecules arrive where the biology is going to grow and the biology grows locally.
What if we grew everything—food, medicine, materials, maybe even complex objects—right where we needed it using distributed biomanufacturing? Can you think of a more compelling vision for an abundant and biotic future?
We met for a walk on campus. As we veered away from Stanford’s Engineering Quad towards the greenhouse to check on Endy’s genetically engineered glowing tobacco plants, I rattled off various ideas about synthetic biology. At a certain point, I mentioned the Design, Build, Test, Learn (DBTL) loop.
DBTL is a succinct meta-level description of engineering. First, Design a system, and then Build a prototype. When the prototype inevitably fails upon Testing, Learn from the failure modes and return to the Design phase. In bioengineering, we expect to run countless Test cycles in the lab before anything works.
Endy sighed and shook his head. He told me he could distinctly remember the slide from a presentation nearly two decades ago that played a central role in sparking the DBTL craze. In that same slide deck, there were other ideas about bioengineering that didn’t stick in nearly the same way. We need:
Standardized parts. Again, how can we do reliable engineering without understanding our basic building blocks?
New abstractions. What is the equivalent of Verilog for biology?
Decoupling of design and fabrication. We need to separate the designers from the builders—like the divide between architecture and construction—to collaborate more efficiently.

These individual pieces comprise a scaleable workflow for engineering living systems. The point isn’t to continually cycle between measurement and design. We should mature into an engineering discipline capable of reliably implementing complex, multi-component designs—even the first time through the workflow.
While all of these ideas have had various degrees of success in making their way into the world, the cyclical nature of this workflow has taken on an outsized role. As a communicator, you can’t entirely control which messages stick, but Endy expressed some regret at this outcome. In a way, we’ve rationalized our lack of progress on part standardization and effective abstractions by convincing ourselves that our trial-and-error processes are just the natural DBTL cycle we’ll always be stuck with.
You may be surprised by this—I certainly was. One of the prevailing views in synthetic biology is that the path forward is to massively scale up each step of the Design, Build, Test, Learn process to overcome biological complexity by brute force. Modern technologies for multiplexed DNA synthesis should be combined with machine learning to engineer new systems through successive rounds of large-scale experimental testing. George Church, one of the pioneers of this paradigm, has referred to this as “ML squared” (Multiplexed Libraries and Machine Learning), and I’ve described it as the 4-S model.
What does this look like in practice?
One of the recent studies I covered provides an excellent example of this approach. A group of synthetic biologists from Rice proposed a new system for designing genetic circuits. They aimed to optimize a drug-inducible gene circuit called synTF:

A gene (eGFP) turns on when a particular molecule is present (4-OHT). Even a simple circuit requires several DNA building blocks. Troubleshooting this type of multi-component circuit is hard. Instead of manually testing design combinations based on intuition, the Rice team screened over 100,000 circuits in a single experiment.

This is the scale that is now possible with modern genomics. Testing this many circuits opens the door to Machine Learning—rather than bespoke Learning via trial and error. Instead of mechanistically understanding how to compose genetic parts, we can generate enough data to learn a computational model of the relationship between DNA composition and function. Essentially, this approach is DBTL on steroids.
The new paper from the Endy Lab, Engineering tRNA abundances for synthetic cellular systems, pursues an orthogonal approach to cellular engineering. All engineering took place in a completely defined synthetic system. Design decisions were made based on a physics-based Computer-Aided Design (CAD) framework, with the goal of working as expected the first time around. Design, Build, Work.
Alright, let’s break down what this means.
Since the 1960s, we’ve refined approaches for extracting the machinery necessary for protein production from cells for use in more controlled chemical reactions. This paradigm, called cell-free protein synthesis, has a wide range of uses.
In reality, the name is a bit of a misnomer. Cell-free technologies use a cell—just one that has been ripped open and sifted through for specific parts. The cell extract is typically a crude mixture.
In 2001, researchers in Japan created a purified system for protein production with the smallest possible set of components. This technology, called PURE, consists of 108 molecules. Each molecule is present in a defined concentration. PURE is an entirely controlled technology for the last step of the central dogma: mRNA to protein.
Because of its precision, PURE is widely used by researchers working to build totally synthetic minimal cells. This was the system that the Endy Lab chose as the basis for their physics-based modeling efforts.
What laws of physics govern the inner world of cells?
Cells are crowded capsules full of biomolecules suspended in solution. Processes such as translation have specific chemical kinetics—the rate at which molecular reactions occur—but the molecules also have volume in space and are constantly moving around. In previous work, the Endy and Zia labs collaborated to build a model that accounted for both the chemistry and the physics of translation elongation.

In their own words:
In our framework, we explicitly represent the transport dynamics of individual biomolecules as they physically interact and chemically react, with nanometer and nanosecond resolution, to simulate processes spanning minutes in time. A key aspect of our approach is the robust modeling of Brownian motion and colloidal-scale particle interactions such that these molecules undergo the inertialess physical encounters appropriate to the colloidal regime.
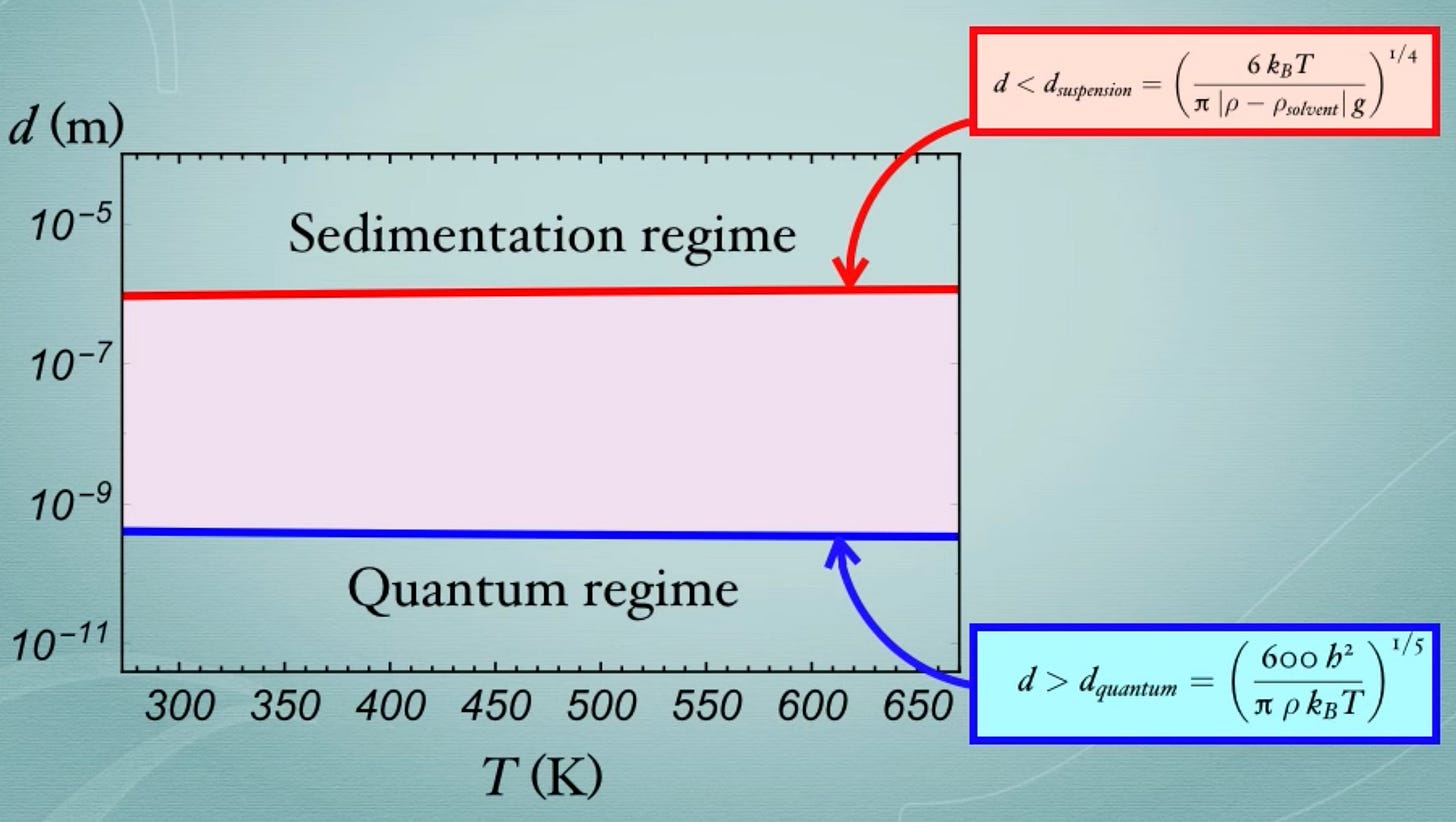
What is the colloidal regime?
Particles of different sizes behave differently. The behavior of tiny particles is mainly dictated by the laws of quantum mechanics. Bigger particles crash out of solution and form sediment. In between these ranges is a Goldilocks zone called the colloidal scale.
Cellular particles exist in this zone. This framework takes that into account and simulates the motion of each particle within the translation elongation reaction accordingly. Physical simulation of cellular systems isn’t new—the tool they adapted was first introduced in 2004—but the focus on accurately modeling colloidal-scale interactions introduced a new level of resolution and predictive power.
This model was the primary product of Akshay Maheshwari’s Ph.D. at Stanford—it took six years of hard work to build.
Now, let’s turn our attention back to the new results. This new study integrates these different pieces together into a predictive engineering system. All of the work happens using PURE, and the system's behavior is modeled using the colloidal physics engine.
What biological process is being engineered?
The rate of protein synthesis. Here’s the problem statement: an engineer asks for a specific rate of protein synthesis using PURE. This simulation framework should reliably spit out a set of transfer RNA (tRNA) abundances that would reliably produce that rate. Importantly, this set of abundances should work the first time around.
Again, translation is a physical process. One of the findings of the initial colloidal modeling work was that the physical transport of complexes around the cell account for ~80% of the latency of translation. Engineering the abundance of tRNAs should reliably control the rate of the reaction.
They built a design tool called Colloidal Dynamics CAD (CD-CAD) to achieve this. CD-CAD used a genetic algorithm—a simple algorithm that selects the fittest outputs of a model over successive rounds of optimization—running on top of their colloidal physics engine to generate the tRNA abundance distributions.
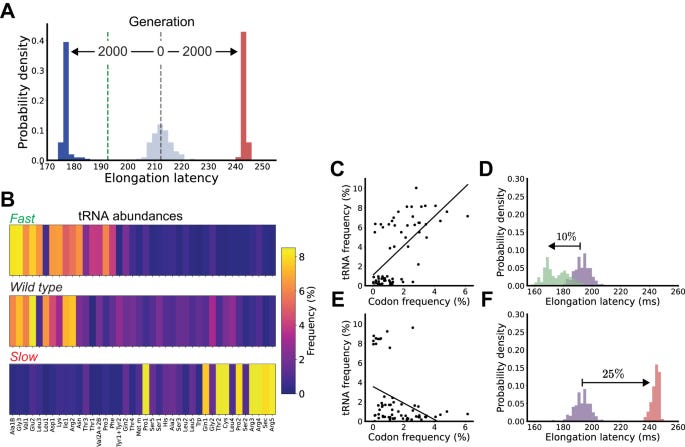
It worked!
Using their CAD tool, they could predictably tune the elongation latency of protein synthesis.
Finally, they also implemented a system for producing any specified distribution of tRNA within PURE, called Tunable Implementation of Nucleic Acids (TINA). Here’s what it all looks like together:

Alright… minimal cell-free systems, colloidal physics, tRNA abundance distributions… technical stuff. Oof! What can we learn from this technology about the future of cell engineering?
Completely specified cells
One of the primary tensions in modern science—especially in disciplines studying emergent systems like biology—is whether we should discover principles that humans can understand or accept that some phenomena will only be understood by machines.
In 2008, Chris Anderson, the editor-in-chief of Wired, argued for the machine-centric scientific approach. In The End of Theory: The Data Deluge Makes the Scientific Method Obsolete, Anderson described a future in which we primarily describe complex natural phenomena in terms of statistical correlations we find in Internet-scale data sets. He wrote,
In short, the more we learn about biology, the further we find ourselves from a model that can explain it.
There is now a better way. Petabytes allow us to say: "Correlation is enough." We can stop looking for models. We can analyze the data without hypotheses about what it might show. We can throw the numbers into the biggest computing clusters the world has ever seen and let statistical algorithms find patterns where science cannot.
At the time, scientists primarily rolled their eyes. Sure, Wired, let’s throw out the scientific method. Fifteen years later, I would argue that this view is surprisingly close to consensus.
As the machine learning revolution sweeps through more domains of science, we’re still looking for models; but we don’t expect to understand them anymore.3 It’s the Bitter Lesson, as Rich Sutton would put it. With more data and computation, search and learning algorithms are what scale—not human-knowledge-based methods.
Doesn’t this describe the high-throughput approach to synthetic biology? It’s way too hard to completely model every interaction between genetic components, so we should design circuits via large-scale parallel DNA synthesis and machine learning.
First principles physics-based modeling is not in vogue. Just this week, the AI-first drug discovery company Recursion Pharmaceuticals announced a giant new in silico drug-target interaction screen, saying they were “able to achieve what would have otherwise taken 100,000 years to compute with physics-based methods.”
But isn’t there something to be said for developing completely defined engineering platforms that we can simulate with molecular resolution? What if we didn’t need to make 2.8 quadrillion predictions but could punch our cellular designs into a physics engine, grab a coffee while the numbers crunched, and come back to boot up a working synthetic cell in the lab?
This study showed that a sliver of this is possible—we can predictably tune protein synthesis rates. Imagine gradually expanding the scope of what we can routinely fabricate and simulate. Not just protein synthesis, but controlled enzymatic DNA synthesis. Completely defined membrane formation.4
In other words, a fully described cell—a human-understandable UNIX-like operating system for cellular programming—that is chemically specified and reliably modeled by physics.
To be clear, I’m a huge fan of data-driven biology. This is why I wanted to explore this new paper—it’s a valuable exercise in challenging assumptions. CD-CAD “did not require feedback or parameterization from our experiments.” It was not a learning algorithm. Learning algorithms incorporate as little bias and structure as possible—everything is learned from parameterization.
What might be possible between these two extremes?
Could we make cell-scale physics simulations faster by learning models of particle interactions? Could we incorporate meaningful physical constraints and parameters into our ML architectures to train them with less data? What tools from the rapidly expanding world of physics-based deep learning could have the most significant impact on bioengineering?
I have more questions than answers here. We’re talking about the frontier of cell programming. But ultimately, we all want to get to the same place. When the inspiration strikes for a new piece of cellular software, we want to be able to…
Design, build, work.
Thanks for reading this essay about physics-based cell engineering. Thanks to Drew Endy and Jon Calles for valuable feedback during the writing process.
If you don’t want to miss upcoming essays, you should consider subscribing for free to have them delivered to your inbox:
Until next time! 🧬
The system was Computer-Aided Three-dimensional Interactive Application (CATIA), originally developed by the French. From Larry Olson, the Director of Information Systems for Boeing Commercial Group, “Hundred percent digital design was a real paradigm shift. The 777 was completed with such precision that it was the first Boeing jet that didn’t need its kinks worked out on an expensive physical mock-up plane."
It’s worth noting that this estimate has decreased over time. The current number of truly unknown genes may be closer to 80.
One fascinating branch of science is ML interpretability, which aims to develop tools to decipher the patterns models are learning to make predictions. People view this field as an essential part of AI safety research, but it is also happening in computational biology, as I’ve previously written about.
To explore more work in this direction from the Endy Lab, check out the preprint describing the “Pureiodic Table.” It represents an attempt to map out “which life-essential functions can work together in remaking one another and what functions remain to be remade.”






This is such a cool example of how to incorporate domain-specific, interdisciplinary knowledge in a tractable way! I also love the questions at the end - incorporating physics constraints is something physicists too are working on with their own algorithms and also helps with the interpretability of the algorithm - really cool to see these principles applied in a cell biology context
Wow, an amazing future ahead!