DNA Ticker Tape
A new DNA-based molecular recording system
Introduction
What is the ideal measurement instrument? I think that the platonic measurement device might look a lot like virtual reality. Imagine being able to conjure arbitrary worlds just like our own, and being able to perform any perturbation you’d like in them. Importantly, imagine being a detached and unobtrusive observer, capable of zooming in and out to any desired scale to examine phenomena as they unfold. In a short story called The Plagiarist, the author Hugh Howey explores the implications of this type of technology for science:
If from the entomology department, one might swoop through the night clouds like a bat, virtual sonar picking up invisible bugs to collect. The climatologists would play like gods bored with their food, sitting over the clouds and swirling them with their fingers, taking notes, testing theories. Geneticists would become the size of molecules and be lost in worlds the scope of Mendelian peas, causing mutations.
While I’m not an expert in quantum computation, I don’t expect to be equipped with the “quantum drives” required for this type of experimentation in this century. But this thought experiment does raise some interesting points about what makes a good measurement. Ideally, we want our recording devices to have as little impact on the system that we are observing as possible.
Many techniques in biology, especially those using sequencing, make destructive measurements. The sample is completely destroyed during the recording event.1 In my last post, I highlighted a recent preprint from the Shendure Lab called ENGRAM, which describes a new nondestructive molecular recorder that records events into DNA.
This preprint was posted alongside “A temporally resolved, multiplex molecular recorder based on sequential genome editing” which is an accompanying body of work from the same lab. The lead author of this work was Junhong Choi. This study introduces another new technology called DNA Ticker Tape, which is a system designed for recording molecular events directly inside of cells.
While we may be corporeal beings with limitations on our direct sensory systems imposed by evolution, I think that it is pretty ingenious that we are beginning to design nanoscale systems to record events at the molecular level as they occur in a nondestructive way.
Key Advances
The core concept of this paper is that it is possible to link events that occur inside of a cell to a downstream recorder that stores information in DNA. This type of futuristic instrument is called a DNA memory device, which the authors define as an “engineered system for digitally recording molecular events through permanent changes to a cell’s genome that can be read out in post hoc fashion.”
Many researchers are beginning to explore new designs for this type of system, each coming with benefits and trade-offs. Some of the limitations of current DNA memory devices include being limited in the number of signals that can be recorded, as well as in the toxicity of the recording event.2 Another key technical challenge is that currently most devices randomly write events to unordered sites in the genome. This makes it very difficult to infer the order of the events being recorded.
The new technology in this paper, DNA Ticker Tape, was designed to address some of these limitations. This device takes inspiration from the way that hard disk drives work. In these hardware systems, a “write head” moves sequentially along a magnetic surface, storing binary data bits as it moves along.

Here, inside the world of the cell, the authors swap out the magnetic recording layer for “a tandem array of partial CRISPR-Cas9 target sites” and the writing device for prime-editing guide RNAs (pegRNAs) that are expressed when specific cellular events occur.
In order to achieve sequential recording, the tape begins with all of the CRISPR-Cas9 target sites in the array made inactive by a 5’ truncation except for the first one.3 Each time an edit is made, the insertion records the cellular signal and activates the next target site in the array by restoring its 5’ sequence. This acts like a write head, moving the next location open for recording one position further along.
In this way, molecular events can be sequentially recorded into DNA as they occur, and can be read after the fact using sequencing.
Results
There are tons of engineering results in this study as the team worked to characterize and optimize this system, and several compelling demonstrations. One of the demonstrations shows how DNA Ticker Tape can be used to record cell lineages during development, which has direct biological relevance. Here, I’m going to focus on the demonstration that shows the amount of fun this lab seems to have while making incredible technology.
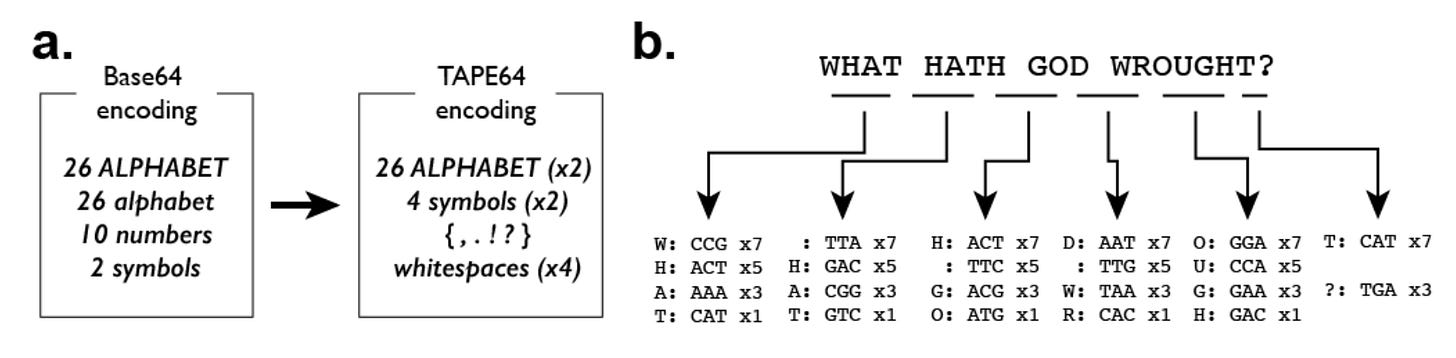
In order to test DNA Ticker Tape, they encoded ordered messages into DNA and attempted to decode them. To do this, they adapted the Base64 encoding scheme that is commonly used to encode text into binary for digital computers, and created TAPE64, mapping the 6-bit binaries that represent characters to the 64 possible 3-mer combinations of DNA bases.
Next, they took three different messages and broke them up into chunks of four characters. Within a given chunk of four characters, the order was encoded by a ratio of 7:5:3:1 for the transfection.
Above, we can see the encoding scheme and an example of how it works for one of the messages they encoded. The message “WHAT HATH GOD WROUGHT?” was selected because it was the first long-distance message transmitted by Morse code in 1844.
In order to decode the message, they first sequenced the array and detected all insertion events. Next they used an algorithm that they established for deriving the ordering of the events. This process consists of analyzing the bigram transition matrix seen above on the left, which can be used to identify the order by determining the frequency at which one of the codes came after the other. Finally, “Within each set of characters inferred to have been co-transfected, ordering was based on corrected unigram counts (middle), resulting in the final decoded message (right).”
They also encoded and decoded two other messages:
“MR. WATSON, COME HERE!”, the first message transmitted by telephone in 1876; and (3) “BOUND FOREVER, DNA”, a translation of a lyric from the 2017 song DNA by the K-pop music group BTS.”
And we can see how they did:
In this creative demonstration, we can see the fidelity with which DNA Ticker Tape can be used to record and decode complex and ordered sets of signals.
Final Thoughts
One of things that I love the most about genetics is that DNA is a beautiful example of information manifested in physical form. In computers, information is represented in binary bits instead of DNA bases. As sequencing has progressed, computation has become an essential aspect of biological research. But united by information theory, these fields blend in surprising ways outside of just data analysis. For example, computer scientists have discovered new algorithms in nature that have improved digital computing systems.
Here, in “A temporally resolved, multiplex molecular recorder based on sequential genome editing” from the Shendure Lab, we see another type of blurring between computer science and genetics. This new technology, DNA Ticker Tape, is a technique for treating the genome like a hard disk drive for recording complex cellular events. It is fun to imagine all sorts of crazy things that this could enable in the future. Could engineered cells with molecular recorders store information about our cellular health, to be later read out and integrated into our medical reports?
As scientists, we may not yet be able to zoom around as disembodied observers in the virtual worlds imagined by Hugh Howey, but we may be able to coax cells into keeping a record of their events for us using molecular recorders. I think that is pretty cool.
If you’ve enjoyed this post and don’t want to miss the next one, you can sign up to have them automatically delivered to your inbox:
Until next time! 🧬
One example of a nondestructive technique is live-cell imaging, where processes are observed as they unfold using microscopy. This type of technique is not always possible, and is not as scaleable as sequencing-based techniques in terms of the number of measurements being made or factors being detected.
For example, a cell can tolerate only a limited number of double-stranded breaks (DSBs).
A quick note here on DNA directionality: there is a chemical convention in the reading/writing of DNA sequences. The 5’ end is the side with a phosphate group on the 5’ carbon, and the 3’ end is the side with an -OH group on the 3’ carbon. DNA is read from the 5’ end to the 3’ end. For more, read here.