Cost-effective age predictions
A new technology called TIME-seq offers the promise of high-throughput epigenetic age prediction
Unfortunately, because aging is so common and natural, we tend to think of it as destiny or something we should accept.
- David A. Sinclair
Introduction
Historically, the most common approach to biomedical research has been to target individual diseases. Our current research paradigm consists of building detailed models of each disease, and tailoring therapeutics specifically towards viable targets in each system independently. A growing faction of researchers are asking the following question: what if there is an underlying phenomenon that is responsible for driving nearly all of human diseases?
The argument from these researchers is that there is such a phenomenon, and that it is aging. One of the core difficulties within the field of aging research is that it can be difficult to achieve consensus on what aging even is, but there is a crucial empirical observation that we are all naturally accustomed to: the longer that an organism lives, nearly all diseases become more likely.
Although the molecular and genetic basis of aging has been studied for decades and organizations such as the SENS Research Foundation have been in existence since the 2000s, there has been a new wave of interesting in aging science in recent years. There are clears signs that this research has become more of a mainstream goal of biotechnology, such as the establishment of a venture fund entirely dedicated to longevity in 2011, and Google’s launch of their anti-aging company Calico Labs in 20131.
Even more recently, the news that several billionaires including Jeff Bezos are investing in a new anti-aging company called Altos Labs made a big splash in the biotech community. This amount of hype has been compounded even further by the newfound interest in longevity of several prominent crypto billionaires such as Vitalik Buterin, who donated around $336 million dollars to anti-aging research.
The influx of an enormous amount of cash and interest into a field riddled with arguments over how to even define itself or effectively measure outcomes can make it challenging to evaluate the amount of scientific progress being made. However, when searching for the signal in the noise, it becomes clear that there are many serious and talented researchers in this space, and that some core principles are beginning to emerge.
For example, it has been established that DNA methylation is one the most accurate biomarkers of aging. This finding has formed the basis for the development of epigenetic clocks, which are models that can be used to predict the age of an organism based on epigenetic measurements. A recent preprint entitled “Ultra-cheap and scalable epigenetic age predictions with TIME-Seq” has recently described a new sequencing technology that holds the promise of greatly decreasing the cost of the measurements needed by epigenetic clocks.
This work is from the Sinclair Lab at Harvard, which has been one of the most prominent academic groups in the field of aging and longevity research. This study was led by the graduate student Patrick Griffin.
Key Advances
The fundamental premise of an epigenetic clock is that DNA methylation is highly predictive of an organism’s age. As input, these models take the methylation levels of CpG sites in the genome, and use them to predict age.

The predictive signal that is observed by these models is that there is an increase in erroneous methylation: sites that were supposed to be methylated no longer are, and previously unmethylated become methylated. This general loss of epigenetic information is one of the most accurate biomarkers of aging, and a lot of work is being put into making them even more accurate. This preprint set out to tackle a different problem, saying “while much focus has been put on developing more accurate clocks, clocks in different organisms, or clocks trained to predict aging- adjacent phenotypes, very little work has been done to make the use of epigenetic clocks more experimentally tractable.”
So what has been the state-of-the-art for making the experimental measurements necessary to make epigenetic clock predictions? There have primarily been two assays for measuring CpG methylation levels: 1) methylation microarrays, and 2) Reduced Representation Bisulfite Sequencing (RRBS).2 The primary drawbacks of these established assays are the high cost associated with each measurement, as well as the extensive sample preparation required. These aspects make it challenging to scale the number of measurements collected.
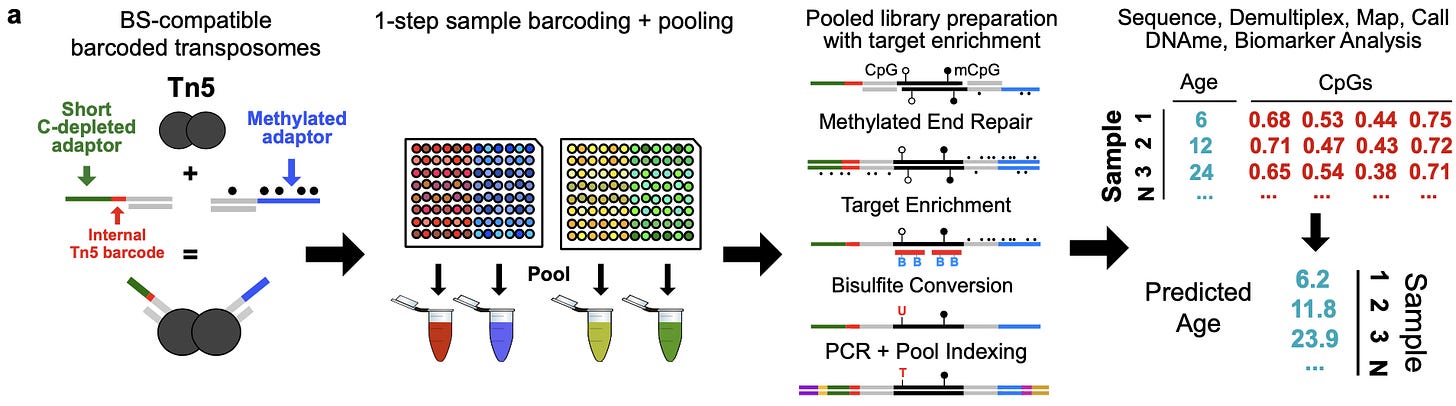
To address this scaling problem, the authors of this study developed a new sequencing technology called Tagmentation-based Indexing for Methylation Sequencing (TIME-Seq):
Similar to other high-throughput sequencing technologies, they used Tn5 transposomes, which are enzymes that can be used to cut up DNA while inserting adapter sequences in a single reaction. Here, the TIME-seq adaptors that get integrated by Tn5 are very short sequences designed for efficiently enriching CpG methylated regions. The resulting fragments with the necessary adapters can be amplified and sequenced. After the pooling step, enrichment is performed, as well as a methylated end repair step. Finally, the sequenced reads are mapped back to individual samples based on the sequencing adaptors that were introduced, and the final set of measurements consists of “a matrix of methylation values for CpGs in each sample.”
Results
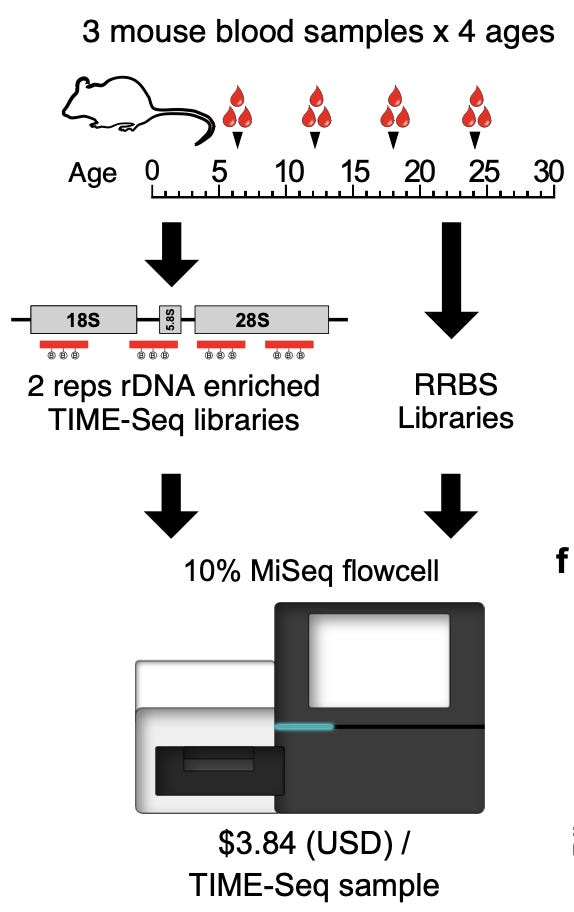
In order to evaluate the accuracy of their new method, the authors first ran a small pilot study comparing TIME-seq (with two replicates) to RRBS libraries as a baseline:
They found that they could effectively demultiplex their pooled libraries, and had high correlation in values between replicates. They also saw good read coverage along their targeted CpG loci with less than 600,000 reads, which meant that their enrichment strategy was working:

With their technique seemingly performing as expected, they generated TIME-seq data for a larger dataset of 191 mouse blood DNA samples, in order to train new epigenetic clocks entirely from new measurements. They trained both an rDNA enriched clock, as well as an epigenetic clock on non-repetitive loci of “CpGs that were previously reported to have high age-correlation in mouse blood.” Like other epigenetic clocks, these models were robust predictors of age:
The key take-home message here is that this sequencing technology can be used to train highly accurate epigenetic clocks like other measurement approaches, but at a fraction of the cost. They wondered if they could take this even further by using a recent probabilistic method called scAge developed for making epigenetic age predictions from single-cell measurements.
The rationale was that they could doing shallower sequencing with TIME-seq (making the cost lower), and that the result would resemble the more sparse data collected from single-cell techniques. They were able to generate effective measurements with shallowing sequencing for $1.85 a sample, which “decreases the cost of age prediction by more than 2 orders of magnitude.”
There are more results worth taking a look at in this paper than I will cover here. They assessed whether or not TIME-seq could detect reliable aging interventions such as caloric restriction. They also evaluated TIME-seq performance on human data, and found that it will be necessary to do more work to refine epigenetic clock models in this context. I find it hard to imagine that improved performance in human samples will be a result that we have to wait too long to see.
Final Thoughts
Aging and longevity research offers the enticing concept that there is a core aspect of physiology which if targeted could prevent most (if not all) diseases. As biotechnology ramps up and more high-profile successes such as the COVID-19 vaccine touch more aspects of our lives, hopes and ambitions for what is possible will continue to grow.3 Biotechnology is being infused with large new sources of capital and interest, and longevity is one of the areas with the highest amount of excitement and intrigue.
Within the eye of this storm, many scientists and engineers are continuing to accrue solid evidence and build important new tools. This new work from the Sinclair Lab has done the pragmatic and important work of building on the stable foundation of epigenetic clocks, and developing a new sequencing approach that makes their construction and predictions much more experimentally tractable. With a low-cost aging biomarker, it may be possible to do new types of studies that were previously completely infeasible, such as large scale clinical trials, or even population-scale studies of aging with epigenetic predictions.
Thanks for reading this highlight of “Ultra-cheap and scalable epigenetic age predictions with TIME-Seq”. If you’ve enjoyed this research highlight, you should consider subscribing so that the next post will arrive in your email inbox:
Until next time! 🧬
After Google’s internal restructuring in 2015, Calico is now an Alphabet company.
These are both fairly mature technologies. Illumina has products for both methylation microarrays, and methylation sequencing.
It is The Century of Biology after all! This train is only leaving the station.