Alignment-free sequencing
A computational pipeline for single-cell sequencing without a reference assembly
Overview
So far since I’ve started writing this newsletter, I’ve spent a decent amount of time talking about the centrality and importance of reference genomes. I’ve also shared my conversation with one of the current pioneers of genome assembly, where we talked about some of the technical complexity involved in creating a new assembly.
In my own work, I’m currently a member of the team building the next version of JBrowse, one of the most popular genome browsers used by model organism communities. I find this to be an especially exciting project because at times these crucial communities can feel underfunded and underserved relative to the human genome research community.
An even further observation can be made: outside of the human reference genome and the model organism communities that we do fund and support, what about the rest of life on Earth?
In my highlight of VeloViz, I talked about the awesome power and rise to prominence of single-cell genomics:
As the cost of next-generation sequencing dropped exponentially after the completion of the Human Genome Project, it became possible to move from sequencing cells in bulk to collecting measurements from individual cells in a sample. This change in resolution is essential for capturing dynamic processes in complex tissue microenvironments.
However, the exciting new technology of single-cell RNA sequencing (scRNA-seq) is largely reliant on the existence of a reference genome to which the new sequencing reads can be aligned to. Unfortunately, that rules out 99.9% of organisms!
In a promising new preprint entitled “Single-cell transcriptomics for the 99.9% of species without reference genomes” a team led by Olga Botvinnik set out to change this. This work proposes a new computational pipeline called Kmermaid that relies on the power of k-mers in an attempt to obviate the need for a reference genome when using scRNA-seq.
Key Advances
At a high level, an experiment using scRNA-seq generates unaligned reads. These are strings of sequences of transcripts that have been measured in the sample, but haven’t been placed on the reference genome to determine what annotated transcripts they actually correspond to. The process of performing this computation is called alignment, and it is a hard problem.
With aligned reads, you can quantify the abundance of transcripts for different genes in a sample, and derive an expression profile for a given cell. These expression profiles can be used to compare new data to existing atlases of cell expression and label cells based on their type.
This is the fundamental goal of this new method: to predict cell type based on scRNA-seq reads. Currently, you can’t do this without a reference genome. Even when there is a reference genome, current approaches for comparing transcriptomes across species in the past have thrown out a lot of data:
At each step in the workflow of propagating labels to cells from a different species, there is data loss. First, a large fraction of the reads simply won’t align to the reference genome.1 After this, data from unannotated regions is also chucked out. The last narrowing step is to remove any data from genes that aren’t actually orthologs2.
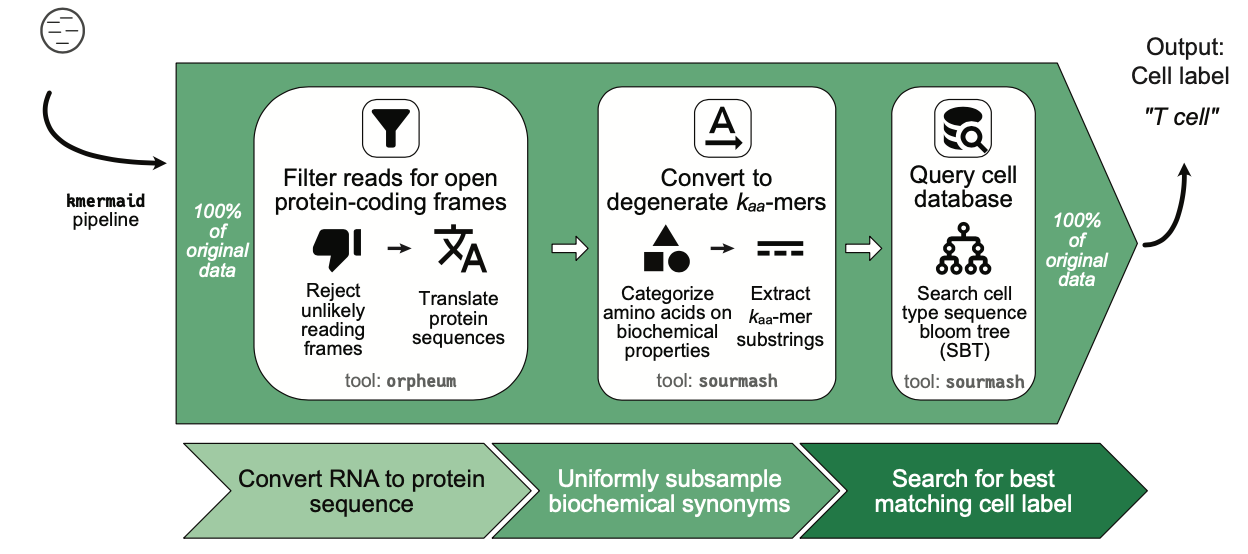
Botvinnik et al. set out to create a less wasteful approach. To do so, they used k-mers, which are one of the workhorses of modern bioinformatics and genomics3. They provide a great visual overview of the new pipeline:
The first step of the pipeline uses a new Python tool the authors built called orpheum to process the reads into amino acid translation frames, because “protein sequences are more evolutionarily conserved than the underlying DNA.”
Next, the pipeline maps the translation frames into a down-sampled k-mer space introduced by Sourmash from the DIB Lab run by C. Titus Brown (also an author on this study too!). This is a cool and efficient technique, where the subsampling serves to “compress data into a smaller form factor, but faithfully represent the underlying data as if all k-mers were present.”
The last step is to use these k-mer representations to search in a database of expression profiles for common cell types to make the final prediction. This constitutes an exciting new paradigm for alignment-free cross-species prediction of cell types that throws out far less data!
A key deliverable of this work is the development of an automated computational pipeline to carry out this workflow for new data. Tangentially, I’m really excited about the increasing adoption and maturity of workflow systems for bioinformatics and genomics. Often, new methods like this one constitute a complex sequence of steps that use some new tools/code and some existing tools/code to transform input data into a desired output. Having standardized tools to build and share these types of workflows is a big win for reproducible science.4
Results
So how well does this new k-mer based cell type prediction pipeline work? The main dataset for evaluation consisted of annotated scRNA-seq from the lung tissue of three species: Chinese horseshoe bat, mouse, and human.
The way that they evaluated accuracy was to see how accurately they could propagate the set of annotations from the mouse cells across all three species. Here is how the data looked for cell compartments. Briefly,
Cell types can be aggregated by their broad functional categories or cell lineage, such as grouping B cells and T cells into a common lymphoid compartment. In this work, we refer to five cellular compartments: endothelial, epithelial, lymphoid, myeloid, and stromal.
Awesome! Their approach (Dayhoff on the right) was able to fairly stably propagate the labels of what compartment the cells belong to across species. At a more granular level, the specific cell type propagation was less accurate, which the authors were able to attribute to subtle misclassifications within the broader categories of compartments with this method.
Ultimately, being able to successfully propagate cell compartment labels across species without any alignment step is a really impressive result, and already opens up new unexplored territory for many biologists. I’m excited to see how this new approach evolves over the years, and whether the cell type propagation performance comes to resemble that of the compartment prediction.
Final Thoughts
It is important to remember just how much incredible biological variation exists within the Earth’s ecosystem that is not represented by reference genomes. This matters for several reasons. First, as scientists and curious explorers of the unknown, all of this variation represents an uncharted frontier for discovery. Additionally, this frontier holds the promise of possessing the next breakthrough technology like CRISPR, just waiting to be discovered.
Excitingly, a new research institute called Arcadia Science was recently formed “for the study of emerging research organisms.”5 They describe their mission in the following way:
One of the great missions of humankind is to more deeply understand life around us. To do this, we need to sample life broadly. There are over one trillion species on this planet. Each of these presents an unparalleled opportunity to ask how evolution has solved problems, leveraging the greatest technology we have — biology itself. At Arcadia, we will support this mission by pursuing and sharing discovery-based research across diverse organisms, thereby unlocking the childlike curiosity in us all.
Tools like Kmermaid hold the promise of making the necessary paradigm shift in bioinformatics to begin making our measurement technologies like scRNA-seq more robustly equipped for use across a more diverse range of organisms. In fact, Olga Botvinnik, the lead author of this study, has joined this new institute to continue this important research direction!
I, for one, can’t wait to see what comes next from this institute!
Thanks for reading this highlight of “Single-cell transcriptomics for the 99.9% of species without reference genomes”. If you’ve enjoyed reading this and would be interested in getting a highlight of a new open-access paper in your inbox each Sunday, you should consider subscribing:
That’s all for this week, have a great Sunday! 🧬
Why? “Most animals in the wild are more heterozygous than laboratory-grown animals; thus their reads tend to have a lower rate of alignment due to high variation between the reference genome and individuals, thus reducing the amount of data used in downstream analyses”
An orthologous gene in two species is derived from a common ancestral copy of the gene found in the ancestor of both species.
The k-mer is a substring of length k of a sequence. For example, ATC is a 3-mer of the sequence ATCG. This concept is widely used in bioinformatics. K-mers are are used all over the place, including in my latest paper where we used them to create a metric of specificity for imaging probes.
I’m really excited about the recent birth of new scientific institutions, such as Arcadia. Another example is New Science which is a new non-profit research institute that gives me the impression of a young Cold Spring Harbor crossed with some Y Combinator tech energy. I’ll be really curious to see where the experiment goes. I hope that this represents the beginning of a Cambrian explosion of new research institutes that provide some positive competitive pressure to the existing research model for basic science in academia.