Inferring natural selection
A new computational framework for detecting signals of positive selection
Nothing in biology makes sense except in the light of evolution.
- Theodosius Dobzhansky
Overview
One of the most beautiful (and challenging) aspects of studying biology is that it requires considering processes that unfold over a multitude of scales. Genomes and the proteins they encode are macromolecules that operate under the laws of physics and chemistry. However, understanding cellular and physiological processes often requires abstracting some of these details away and using different tools to model and understand complex systems.
Zooming even further out, biological organisms exist within populations, and these populations are constantly changing according to evolutionary dynamics. One of the disciplines devoted to the quantitative study of these dynamics is population genetics.
My favorite succinct description of population genetics is from Molly Przeworski’s website:
The heritable differences among us reflect accidental changes to the genome that arose in an individual, were shuffled into new combinations by recombination, and persisted in the population, whether by chance or because they were beneficial. Although these processes and their interactions are extremely complicated, they comprise the whole picture—which means we population geneticists are in the somewhat unusual position of knowing what we need to understand. To make sense of the variation within species—whether humans or fruit flies—and the differences between species, we need to model and characterize the interactions of mutation, recombination, demography, and natural selection.
This clearly states the research goal of population geneticists. The goal of the field is to effectively model and understand the dynamics of these four forces (mutation, recombination, demography, natural selection).
Excitingly, population genetics has undergone a renaissance due to the combination of two revolutions: the digital revolution and the genomic revolution. The dramatically increased speed and decreased cost of computation has enabled population geneticists to run large-scale computational simulations of theoretical models of evolution that previous generations of researchers could only speculate about on a chalk board. Even better, with the abundance of genomic data, the simulations can be compared to actual data to evaluate the accuracy of the underlying models.
In addition to building and evaluating theoretical models of evolution, one of the primary goals of population genetics is to detect signals of natural selection from genomic data. This is a problem where inference is the key goal. The challenge consists of building a sufficiently accurate and predictive model to be able to identify sites in a genome where evolution is occurring. This type of detection is important for several reasons: it can inform our understanding of the evolutionary past as well as shed light on the mechanistic role of genetic variation as it pertains to health and disease.
As a computational biologist, I have always had an appreciation for population genetics simply because it is really cool. I appreciate the field’s emphasis on modeling and simulation, and often find the papers very satisfying.
One of the lines of research that I have found especially cool and exciting is the intersection of machine learning and population genetics. While this has been studied for a while, it first came on to my radar through the work of Andy Kern and Dan Schrider who were using simulations and supervised machine learning to detect evolutionary selective sweeps in the genome.1 When I first read about this, I thought it was so cool that it was hard to avoid being nerd sniped and resist the urge to drop the project I was working on at the time to study this!

An exciting new preprint entitled “SIA: Selection Inference Using the Ancestral Recombination Graph” from the Siepel Lab at Cold Spring Harbor Laboratory has come out. This study introduces a new methodological advance for this field. Led by Hussein A. Hejase and Ziyi Mo, this work proposes a new method that leverages the power of simulation and ML to detect genomic signals of positive selection.
Key Advances
It’s worth briefly backing up and providing a short primer on positive selection before we look at how to detect it. Populations of organisms can respond to pressures in their environment through the process of adaptive evolution, which “is driven by increases in frequency of alleles that enhance reproductive fitness.”
From a phenotypic perspective, this looks like the change in the distribution of the phenotype under selection within the population that can be seen above. At the level of the genome, the sites containing the alleles under selection are what is increasing in frequency:
So if that is positive selection, what signal will it leave in the genome and how can we detect it? Historically, the approach has been to use theory to devise test statistics that measure changes in allelic frequency caused by selective sweeps. However, as I mentioned earlier, groups have innovated on this problem, bringing new state-of-the-art results by combining summary statistics into more complex models using supervised machine learning or other types of statistical modeling techniques.
This preprint takes a step beyond this formulation, using a richer representation of the underlying evolutionary dynamics than summary statistics: the ancestral recombination graph (ARG). Here is how the authors describe what the ARG is:
The ARG is a complex data structure that summarizes the shared evolutionary history and recombination events that have occurred in a collection of DNA sequences, and therefore contains highly informative features that can potentially be leveraged to make accurate inferences about selection.
Ian Holmes (my boss) provides a good succinct technical description of the ARG: “The ancestral recombination graph is a DAG whose nodes correspond to genotypes and edges correspond to either recombination or coalescence events.”2
With this idea mind, the SIA method becomes more tangible. The method consists of several steps. First, using demographic estimates of the population being studied, computational simulations are used to generate examples of neutral regions and selective sweeps. The ARG is inferred from the simulated data, and processed into numerical features. Those features are used as input for a Recurrent Neural Network (RNN) that was trained to detect selection. Here is what this looks like:
Why does a Recurrent Neural Network make sense? The particular type of RNN that was used was a Long Short-Term Memory (LSTM) network, which is comprised of “cells” that have the ability to keep track of hidden state.

More generally in machine learning, LSTMs are typically used for problems where there is a concept of time, such as time series data, or sequences of video or speech. That was also the motivation here, since evolution happens over time: “SIA uses a Long Short-Term Memory (LSTM) architecture, designed specifically to handle the temporal nature of the feature set. The LSTM unrolls temporally such that the lineage counts at each time point are fed to the network iteratively.”
Between the use of the ARG features and the LSTM architecture, the SIA method contains some interesting new conceptual advances for detecting signals of genomic selection.
Results
This paper is jam packed with results of two primary categories: 1) methodological evaluation, and 2) new inferences made applying SIA to genomic data. We’ll look at one example of each.
One of the first sets of results is the comparison of performance between SIA and comparable methods for detecting sweeps:
Each of these plots is a receiver operating characteristic (ROC) curve, one of the most common plots for evaluating classification performance. The more the curve hugs the top left, the better the classification performance. The plots are ordered horizontally by the selection regime (s) and vertically by the derived allele frequencies for the mutation under selection (f) used for simulation3.
There are some initial observations to make. First, we can empirically see that both a higher s and f value makes for an easier prediction problem, respectively. So, the best results across all models are seen in the bottom right, and the worst in the top left.
Another general observation can be made: “SIA outperformed the other methods across model conditions, with a more pronounced performance advantage for sites under weaker selection and segregating at lower DAFs.” From the evaluation, it appears that by coupling the rich feature set of the ARG values with a LSTM model, SIA has arrived at a more accurate method for detecting selective sweeps.
Being primarily a methodological study, there are many more evaluation experiments and comparisons to other models. However, I want to shift gears and look at an example of detecting sites of selection in the genome.
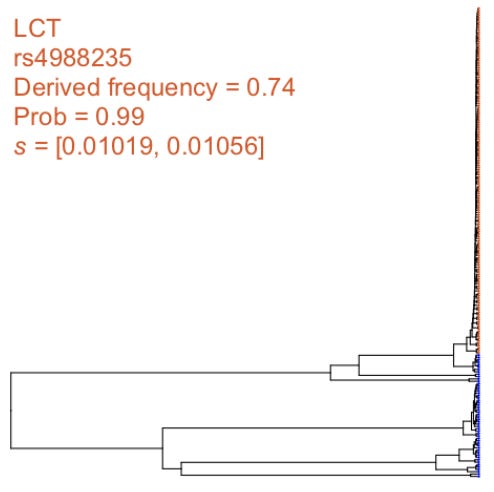
SIA was used to identify selective sweeps and infer selection coefficients in the European (CEU) population of the 1000 Genomes Project dataset. This is an interesting test case, because “These loci included the canonical example of selection at the MCM6 gene, which regulates the neighboring LCT gene and contributes to the lactase persistence trait the ABCC11 gene regulating earwax production, several pigmentation-related genes, as well as genes associated with obesity, diabetes and addiction.”4
SIA picked up a very strong signal for this region: “At this SNP, SIA inferred a sweep probability close to 1 and a selection coefficient greater than 0.01, making this one of the strongest signals of selection in the human genome. A close examination of the local genealogy at this site reveals a clear pattern indicative of a selective sweep — a burst of recent coalescence among the derived lineages (orange taxa are the lineages carrying the derived allele) is clearly visible from the tree.”

In addition to the detection of more canonical sites, SIA was able to clarify, refine, and in some cases provide counter-evidence against a number of sites in the genome that have been hypothesized to be under selection.
Those are all of the results I’m going to highlight here, but there is a lot more interesting data to be found in the paper.
Final Thoughts
As Theodosius Dobzhansky argued in his 1973 essay, “nothing in biology makes sense except in the light of evolution.” When examining how organisms became the way they are, or attempting to understand the functional impact of genetic variation, evolutionary information can be profoundly informative. For this reason, one of the most central problems that population geneticists work on is devising ways to detect these evolutionary signals in genomic data.
This new methodological advance from the Siepel Lab constitutes a powerful new tool in the arsenal of scientists attempting to detect positive selection in genomes. It is an example of the potential promise at the interface of machine learning and population genetics to tackle challenging and longstanding problems.
Thanks for reading this highlight of “SIA: Selection Inference Using the Ancestral Recombination Graph”. If you’ve enjoyed reading this and would be interested in getting a highlight of a new open-access paper in your inbox each Sunday, you should consider subscribing:
That’s all for this week, have a great Fourth of July! 🇺🇸🎇
They wrote a great review of this research entitled Supervised Machine Learning for Population Genetics: A New Paradigm
Before moving on, I’ll make one stylistic note here. I realize that between DAGs, recombination, and coalescence, very few readers will know all of these concepts coming in. In my liberal use of external links, I want to provide a choose your own adventure experience. While I’m going to focus on conceptual overviews of the work I cover in COB and avoid relentlessly defining each new piece of terminology, I hope that the links I provide are useful resources for diving deeper into technical details that you may want to know more about.
The selection regime is a coefficient that measures the difference in relative fitness of the allele under selection. The derived allele frequency measures the percentage at which the allele is present at its site relative to other alleles at that site in the population.
Would you have predicted those alleles as some of the traits most directly under selection?! I think I probably have a hyperactive copy of ABCC11. (Thanks, dad)